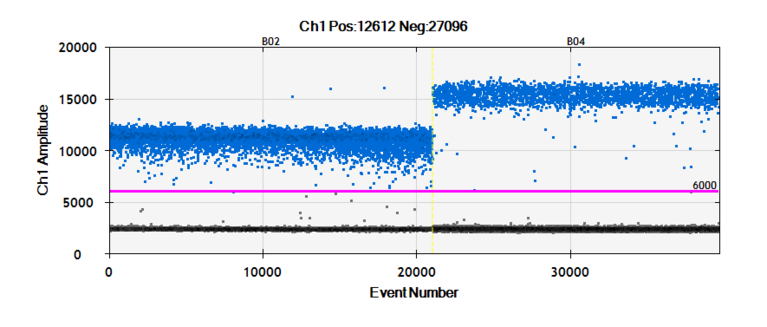
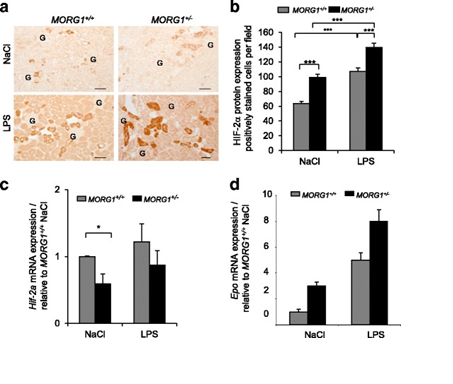
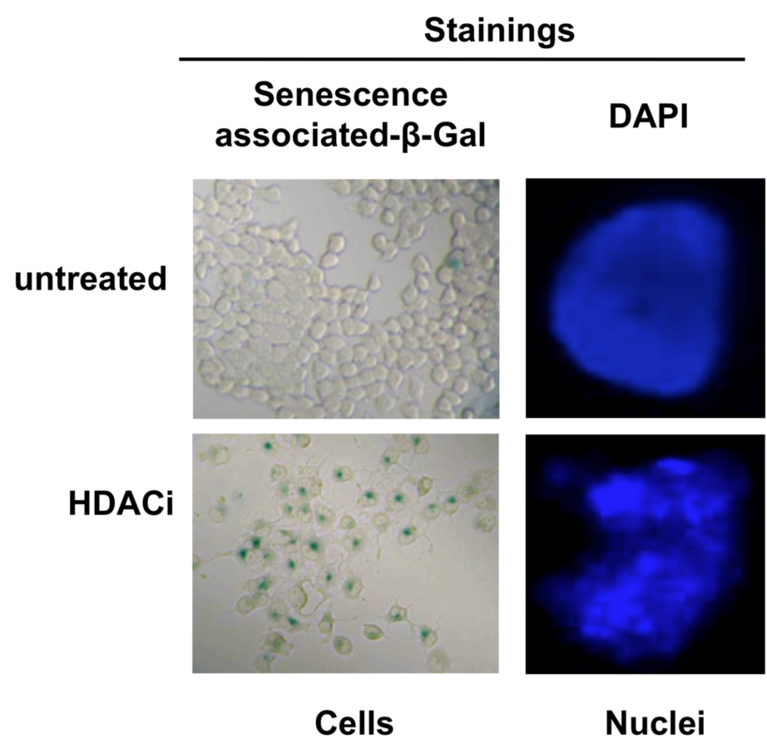
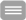Figure 1 a-d: Reduced expression of MORG1 in MORG1+/- heterozygous mice is associated with an elevated renal expression of HIF-2a protein, respectively an increased mRNA levels of the HIF-2a target gene erythropoietin (Epo) in LPS treated mice, compared to the MORG1+/+ wild type endotoxemic mice.
Bondeva T., Schindler C., Schindler K., Wolf, G. MORG1(+/-) mice are protected from histological renal damage and inflammation in a murine model of endotoxemia. BMC Nephrol. 2018 Feb 5; 19 (1):29.