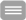A prerequisite for establishment of cellular morphology is the ability of cells to remodel their plasma membrane and to reorganize the actin cytoskeleton in response to external and internal cues. In neurons, actin dynamics and membrane remodelling are indispensable for neuronal network formation, for information processing in the central nervous system and for learning and memory processes. Alterations in dendrite morphology or defects in neuronal development contribute to several neurological and neurodevelopmental disorders, such as autism spectrum disorders, Alzheimer's disease, schizophrenia, Down syndrome and depression.
A major research focus of our groups therefore is to understand how reshaping of the plasma membrane is elicited by proteins, which directly interact with and/or deform the lipid bilayer (membrane shapers), and this is promoted by dynamics of the cortical actin cytoskeleton.
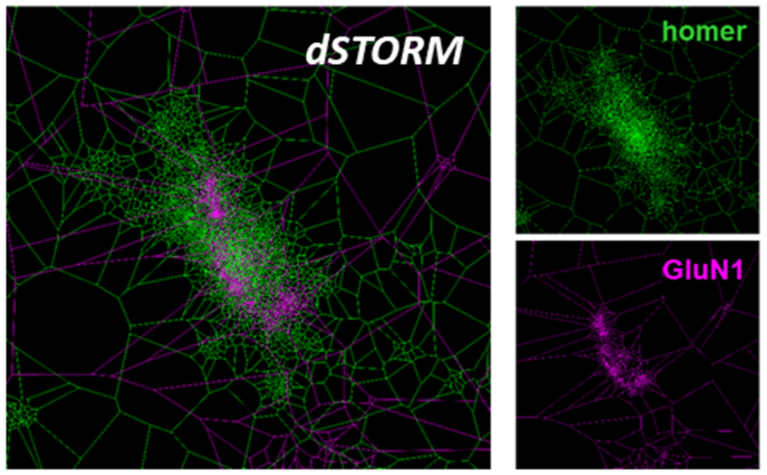
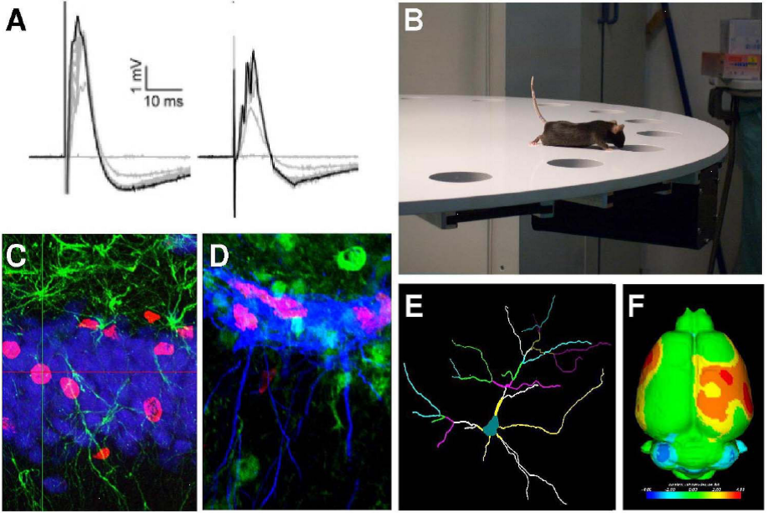
In the summer school, we will apply modern techniques in molecular biology, cell biology and microscopy to study the organization and dynamics of the actin cytoskeleton, effector molecules and Ca2+-signals. Imaging neuronal cell dynamics reveals how structural and functional cellular plasticity is brought about. These processes are indispensable for adaptations of neuronal contacts and reorganizations underlying brain development, regeneration, learning and memory.
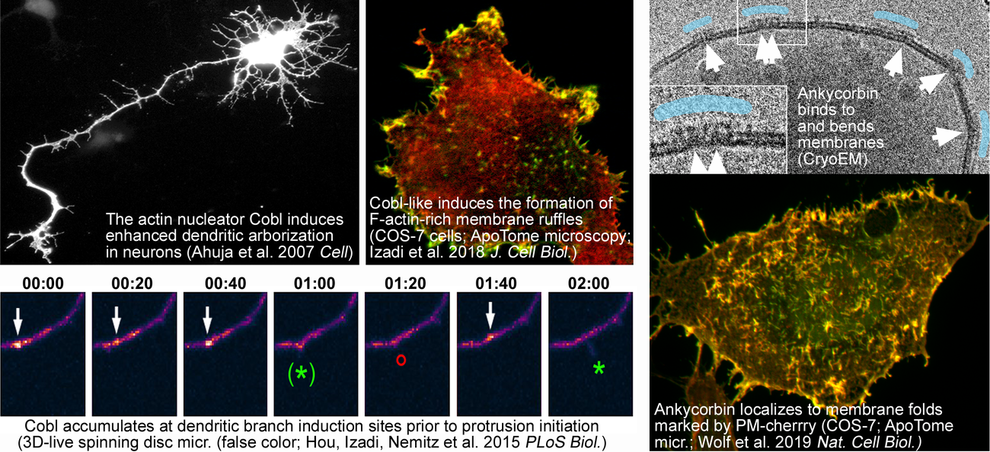
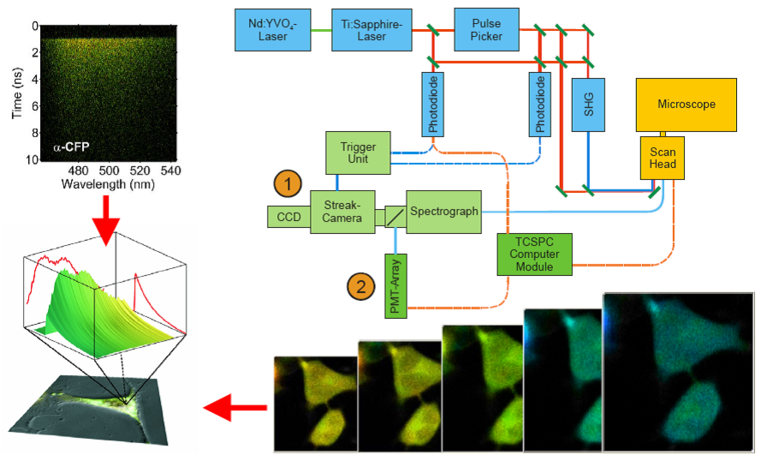
Modern molecular tools will allow for a state-of-the-art analysis of cytoskeletal structures at high resolution. Students will have the opportunity to gain hands-on expertise in live cell imaging using apotome-based confocal microscopy as well as spinning disk microscopy. High-resolution 3D-live analyses will be used to track cellular morphology and proteins implicated in neuronal network formation and nerve cell communication in time and space and to study their dynamics.
The studies in the course of this summer school project will thereby shed light on the fascinating cellular adaptations underlying development, plasticity and information processing/learning in vertebrate brains and on how these functions are derailed in pathological and disease states.